- Article
- Published: 22 February 2021
Genomic insights into the formation of human populations in East Asia
- Chuan-Chao Wang,
- Hui-Yuan Yeh,
- Alexander N. Popov,
- Hu-Qin Zhang,
- Hirofumi Matsumura,
- Kendra Sirak,
- Olivia Cheronet,
- Alexey Kovalev,
- Nadin Rohland,
- Alexander M. Kim,
- Swapan Mallick,
- Rebecca Bernardos,
- Dashtseveg Tumen,
- Jing Zhao,
- Yi-Chang Liu,
- Jiun-Yu Liu,
- Matthew Mah,
- Ke Wang,
- Zhao Zhang,
- Nicole Adamski,
- Nasreen Broomandkhoshbacht,
- Kimberly Callan,
- Francesca Candilio,
- Kellie Sara Duffett Carlson,
- …
- David Reich
Nature volume 591, pages413–419 (2021)Cite this article
- 27k Accesses
- 169 Citations
- 240 Altmetric
- Metricsdetails
출처; Genomic insights into the formation of human populations in East Asia | Nature
Abstract
The deep population history of East Asia remains poorly understood owing to a lack of ancient DNA data and sparse sampling of present-day people1,2. Here we report genome-wide data from 166 East Asian individuals dating to between 6000 BC and AD 1000 and 46 present-day groups. Hunter-gatherers from Japan, the Amur River Basin, and people of Neolithic and Iron Age Taiwan and the Tibetan Plateau are linked by a deeply splitting lineage that probably reflects a coastal migration during the Late Pleistocene epoch. We also follow expansions during the subsequent Holocene epoch from four regions. First, hunter-gatherers from Mongolia and the Amur River Basin have ancestry shared by individuals who speak Mongolic and Tungusic languages, but do not carry ancestry characteristic of farmers from the West Liao River region (around 3000 BC), which contradicts theories that the expansion of these farmers spread the Mongolic and Tungusic proto-languages. Second, farmers from the Yellow River Basin (around 3000 BC) probably spread Sino-Tibetan languages, as their ancestry dispersed both to Tibet—where it forms approximately 84% of the gene pool in some groups—and to the Central Plain, where it has contributed around 59–84% to modern Han Chinese groups. Third, people from Taiwan from around 1300 BC to AD 800 derived approximately 75% of their ancestry from a lineage that is widespread in modern individuals who speak Austronesian, Tai–Kadai and Austroasiatic languages, and that we hypothesize derives from farmers of the Yangtze River Valley. Ancient people from Taiwan also derived about 25% of their ancestry from a northern lineage that is related to, but different from, farmers of the Yellow River Basin, which suggests an additional north-to-south expansion. Fourth, ancestry from Yamnaya Steppe pastoralists arrived in western Mongolia after around 3000 BC but was displaced by previously established lineages even while it persisted in western China, as would be expected if this ancestry was associated with the spread of proto-Tocharian Indo-European languages. Two later gene flows affected western Mongolia: migrants after around 2000 BC with Yamnaya and European farmer ancestry, and episodic influences of later groups with ancestry from Turan.
2017 Feb
Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago - PMC (nih.gov)
Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago
Early Neolithic (~7700-year-old) genetic data from the Russian Far East implies a high level of genetic continuity in this region.Ancient genomes have revolutionized our understanding of Holocene prehistory and, particularly, the Neolithic transition ...
www.ncbi.nlm.nih.gov

RESULTS
Samples, sequencing, and authenticity
To fill this gap in our knowledge about the Neolithic in East Asia, we sequenced to low coverage the genomes of five early Neolithic burials (DevilsGate1, 0.059-fold coverage; DevilsGate2, 0.023-fold coverage; and DevilsGate3, DevilsGate4, and DevilsGate5, <0.001-fold coverage) from a single occupational phase at Devil’s Gate (Chertovy Vorota) Cave in the Primorye Region, Russian Far East, close to the border with China and North Korea (see the Supplementary Materials). This site dates back to 9.4 to 7.2 ka, with the human remains dating to ~7.7 ka, and it includes some of the world’s earliest evidence of ancient textiles (10). The people inhabiting Devil’s Gate were hunter-fisher-gatherers with no evidence of farming; the fibers of wild plants were the main raw material for textile production (10). We focus our analysis on the two samples with the highest sequencing coverage, DevilsGate1 and DevilsGate2, both of which were female. The mitochondrial genome of the individual with higher coverage (DevilsGate1) could be assigned to haplogroup D4; this haplogroup is found in present-day populations in East Asia (11) and has also been found in Jomon skeletons in northern Japan (2). For the other individual (DevilsGate2), only membership to the M branch (to which D4 belongs) could be established. Contamination, estimated from the number of discordant calls in the mitochondrial DNA (mtDNA) sequence, was low {0.87% [95% confidence interval (CI), 0.28 to 2.37%] and 0.59% (95% CI, 0.03 to 3.753%)} on nonconsensus bases at haplogroup-defining positions for DevilsGate1 and DevilsGate2, respectively. Using schmutzi (12) on the higher-coverage genome, DevilsGate1 also gives low contamination levels [1% (95% CI, 0 to 2%); see the Supplementary Materials]. As a further check against the possible confounding effect of contamination, we made sure that our most important analyses [outgroup f3 scores and principal components analysis (PCA)] were qualitatively replicated using only reads showing evidence of postmortem damage (PMD score of at least 3) (13), although these latter results had a high level of noise due to the low coverage (0.005X for DevilsGate1 and 0.001X for DevilsGate2).
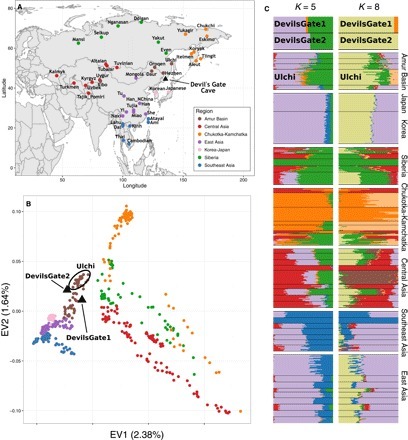
(A) Map of Asia showing the location of Devil’s Gate (black triangle) and of modern populations forming the regional panel of our analysis. (B) Plot of the first two principal components as defined by our regional panel of modern populations from East Asia and central Asia shown on (A), with the two samples from Devil’s Gate (black triangles) projected upon them (18). (C) ADMIXTURE analysis (19) performed on Devil’s Gate and our regional panel, for K = 5 (lowest cross-validation error) and K = 8 (appearance of Devil’s Gate–specific cluster).
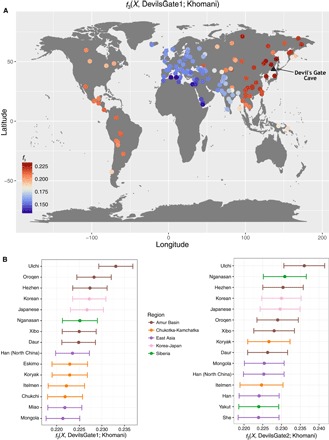
Outgroup f3 measuring shared drift between Devil’s Gate (black triangle shows sampling location) and modern populations with respect to an African outgroup (Khomani). (A) Map of the whole world. (B) Fifteen populations with the highest shared drift with Devil’s Gate, color-coded by regions as in Fig. 1. Error bars represent 1 SE.
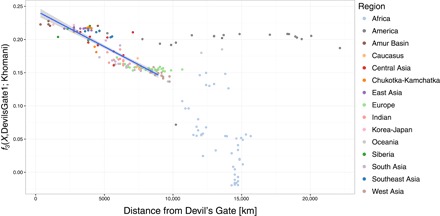
Relationship between outgroup f3(X, Devil’s Gate; Khomani) and distance on land from Devil’s Gate using DevilsGate1 and all single-nucleotide polymorphisms (SNPs). Populations up to 9000 km away from Devil’s Gate were considered when computing correlation. The highest distance considered was chosen to acquire the highest Pearson correlation in steps of 500 km. Best linear fit (r2 = 0.772, F1,108 = 368.4, P < 0.001) is shown as blue line, with 95% CI indicated by the shaded area.
A Dynamic 6,000-Year Genetic History of Eurasia’s Eastern Steppe
출처; A Dynamic 6,000-Year Genetic History of Eurasia’s Eastern Steppe - ScienceDirect
Summary
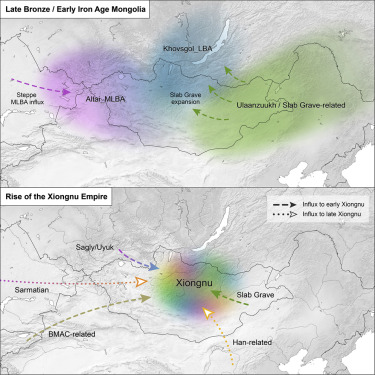
The Eastern Eurasian Steppe was home to historic empires of nomadic pastoralists, including the Xiongnu and the Mongols. However, little is known about the region’s population history. Here, we reveal its dynamic genetic history by analyzing new genome-wide data for 214 ancient individuals spanning 6,000 years. We identify a pastoralist expansion into Mongolia ca. 3000 BCE, and by the Late Bronze Age, Mongolian populations were biogeographically structured into three distinct groups, all practicing dairy pastoralism regardless of ancestry. The Xiongnu emerged from the mixing of these populations and those from surrounding regions. By comparison, the Mongols exhibit much higher eastern Eurasian ancestry, resembling present-day Mongolic-speaking populations. Our results illuminate the complex interplay between genetic, sociopolitical, and cultural changes on the Eastern Steppe.
Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations
Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations - PMC (nih.gov)
Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations
Han Chinese, Japanese and Korean, the three major ethnic groups of East Asia, share many similarities in appearance, language and culture etc., but their genetic relationships, divergence times and subsequent genetic exchanges have not ...
www.ncbi.nlm.nih.gov
We found that the present-day Han Chinese and Japanese have the most recent common ancestor that can be dated back about 3.0~ 3.6 KYA (corresponding to the Shang Dynasty in Chinese history). Korean and northern Han Chinese had frequent communications in ancient time, and the divergence time between the two populations was estimated as ~ 1.2 KYA (corresponding to the later period of Three Kingdoms of Korea, or the Tang Dynasty in China). And Japanese and Korean separated ~ 1.4 KYA, a little earlier than that of Han Chinese and Korean (corresponding to Asuka period in Japan, or in the middle of Three Kingdoms period of Korea).
Posted October 22, 2018.
The population history of northeastern Siberia since the Pleistocene | bioRxiv
The population history of northeastern Siberia since the Pleistocene
Far northeastern Siberia has been occupied by humans for more than 40 thousand years. Yet, owing to a scarcity of early archaeological sites and human remains, its population history and relationship to ancient and modern populations across Eurasia and the
www.biorxiv.org

Reconstructing genetic history of Siberian and Northeastern European populations (cshlp.org)
Reconstructing genetic history of Siberian and Northeastern European populations
Reconstructing genetic history of Siberian and Northeastern European populations Emily H.M. Wong1, Andrey Khrunin2, Larissa Nichols2, Dmitry Pushkarev3, Denis Khokhrin2, Dmitry Verbenko2, Oleg Evgrafov4, James Knowles4, John Novembre5, Svetlana Limborska2
genome.cshlp.org
Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe | PLOS Genetics
Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
Author Summary The history of human populations can be retraced by studying the archaeological and anthropological record, but also by examining the current distribution of genetic markers, such as the maternally inherited mitochondrial DNA. Ancient DNA re
journals.plos.org

Figure 3. Map of genetic distances between modern-day populations of Eurasia and from aUzPo and aBOO.
Genetic distances were computed between 144 modern-day populations geographically delineated across Eurasia (red dots) and the eleven individuals from aUzPo (A) and the 23 individuals from aBOO (B). The colour gradient represents the degree of similarity between the modern and ancient populations, interpolated between sampling points: from ‘green’ for high similarity or small genetic distance to ‘brown’ for low similarity. ‘K’ designates the number of populations used for distance computation and mapping; ‘N’ represents the number of points in the grid used for extrapolation; ‘min’, corresponds to the minimal values respectively of the computed distances between ancient and modern populations.
https://doi.org/10.1371/journal.pgen.1003296.g003

Figure 6Alternative models for the origin of New World founder haplotypes. Numbers in circles on the solid-lined arrows refer to models 1–3 as defined in the text. These illustrative arrows do not denote precisely defined geographic routes. The dotted lines extending into the large circle indicate the possible ancestral homeland of the Kets/Selkups (haplotype 1C-12A) and of the Baikal Region populations (haplotype 1F-11C).
PNAS November 27, 2018 115 (48) E11248-E11255; first published November 5, 2018;
Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe | PNAS
Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe
Since the Bronze Age, pastoralism has been a dominant subsistence mode on the Western steppe, but the origins of this tradition on the Eastern steppe are poorly understood. Here we investigate a putative early pastoralist population in northern Mongolia an
www.pnas.org
Science 29 Jun 2018:
The first horse herders and the impact of early Bronze Age steppe expansions into Asia | Science (sciencemag.org)
The first horse herders and the impact of early Bronze Age steppe expansions into Asia
The Eurasian steppes reach from the Ukraine in Europe to Mongolia and China. Over the past 5000 years, these flat grasslands were thought to be the route for the ebb and flow of migrant humans, their horses, and their languages. de Barros Damgaard et al. p
science.sciencemag.org
First published: 14 June 2018
New genetic evidence of affinities and discontinuities between bronze age Siberian populations - Hollard - 2018 - American Journal of Physical Anthropology - Wiley Online Library
New genetic evidence of affinities and discontinuities between bronze age Siberian populations
Objectives This work focuses on the populations of South Siberia during the Eneolithic and Bronze Age and specifically on the contribution of uniparental lineage and phenotypical data to the questio...
onlinelibrary.wiley.com
Massive migration from the steppe was a source for Indo-European languages in Europe
Published online 2015 Mar 2. doi: 10.1038/nature14317
Massive migration from the steppe was a source for Indo-European languages in Europe - PubMed
We generated genome-wide data from 69 Europeans who lived between 8,000-3,000 years ago by enriching ancient DNA libraries for a target set of almost 400,000 polymorphisms. Enrichment of these positions decreases the sequencing required for genome-wide anc
pubmed.ncbi.nlm.nih.gov
Y Chromosome analysis of prehistoric human populations in the West Liao River Valley, Northeast China
Published online 2013 Sep 30. doi: 10.1186/1471-2148-13-216
Y Chromosome analysis of prehistoric human populations in the West Liao River Valley, Northeast China - BMC Ecology and Evolutio
Background The West Liao River valley in Northeast China is an ecologically diverse region, populated in prehistory by human populations with a wide range of cultures and modes of subsistence. To help understand the human evolutionary history of this regio
bmcecolevol.biomedcentral.com
Genetic characteristics and migration history of a bronze culture population in the West Liao-River valley revealed by ancient DNA